In a previous tutorial, we talked about NumPy arrays and we saw how it makes the process of reading, parsing and performing operations on numeric data a cakewalk. In this tutorial, we will discuss the NumPy loadtxt method that is used to parse data from text files and store them in an n-dimensional NumPy array. Then we can perform all sorts of operations on it that are possible on a NumPy array. np.loadtxt offers a lot of flexibility in the way we read data from a file by specifying options such as the data type of the resulting array, how to distinguish one data entry from the others through delimiters, skipping/including specific rows, etc. We’ll look at each of those ways in the following tutorial.
In a previous tutorial, we talked about NumPy arrays and we saw how it makes the process of reading, parsing and performing operations on numeric data a cakewalk. In this tutorial, we will discuss the NumPy loadtxt method that is used to parse data from text files and store them in an n-dimensional NumPy array. Then we can perform all sorts of operations on it that are possible on a NumPy array. np.loadtxt offers a lot of flexibility in the way we read data from a file by specifying options such as the data type of the resulting array, how to distinguish one data entry from the others through delimiters, skipping/including specific rows, etc. We’ll look at each of those ways in the following tutorial.
Specifying the file path
Let’s look at how we can specify the path of the file from which we want to read data.
We’ll use a sample text file for our code examples, which lists the weights (in kg) and heights (in cm) of 100 individuals, each on a row.
I will use various variants in this file for explaining different features of the loadtxt function.
Let’s begin with the simplest representation of the data in a text file. We have 100 lines (or rows) of data in our text file, each of which comprises 2 floating-point numbers separated by a space.
The first number on each row represents the weight and the second number represents the height of an individual.
Here’s a little glimpse from the file:
110.90 146.03
44.83 211.82
97.13 209.30
105.64 164.21
This file is stored as `weight_height_1.txt`.
Our task is to read the file and parse the data in a way that we can represent in a NumPy array.
We’ll import the NumPy package and call the loadtxt method, passing the file path as the value to the first parameter filePath.
import numpy as np
data = np.loadtxt("./weight_height_1.txt")
Here we are assuming the file is stored at the same location from where our Python code will run (‘./’ represents current directory). If that is not the case, we need to specify the complete path of the file (Ex: “C://Users/John/Desktop/weight_height_1.txt”)
We also need to ensure each row in the file has the same number of values.
The extension of the file can be anything other than .txt as long as the file contains text, we can also pass a generator instead of a file path (more on that later)
The function returns an n-dimensional NumPy array of values found in the text.
Here our text had 100 rows with each row having 2 float values, so the returned object data will be a NumPy array of shape (100, 2) with the float data type.
You can verify this by checking ‘shape’ and ‘dtype’ attribute of the returned data:
print("shape of data:",data.shape)
print("datatype of data:",data.dtype)
Output:

Specifying delimiters
A delimiter is a character or a string of characters that separates individual values on a line.
For example, in our earlier file, we had the values separated by a space, so in that case, the delimiter was a space character (” “).
However, some other files may have a different delimiter, for instance, CSV files generally use comma (“,”) as a delimiter. Another file may have a semicolon as a delimiter.
So we need our data loader to be flexible enough to identify such delimiters in each row and extract the correct values from them.
This can be achieved by passing our delimiter as a parameter to the np.loadtxt function.
Let us consider another file ‘weight_height_2.txt’, it has the same data content as the previous one, but this time the values in each row are separated by a comma:
110.90, 146.03
44.83, 211.82
97.13, 209.30
…
…
We’ll call the np.loadtxt function the same way as before, except that now we pass an additional parameter – ‘delimiter’:
import numpy as np
data = np.loadtxt("./weight_height_2.txt", delimiter = ",")
This function will return the same array as before.
- In the previous section, we did not pass delimiter parameter value because np.loadtxt() expects space “ “ to be the default delimiter
- If the values on each row were separated by a tab, in that case, the delimiter would be specified by using the escape character “\t”
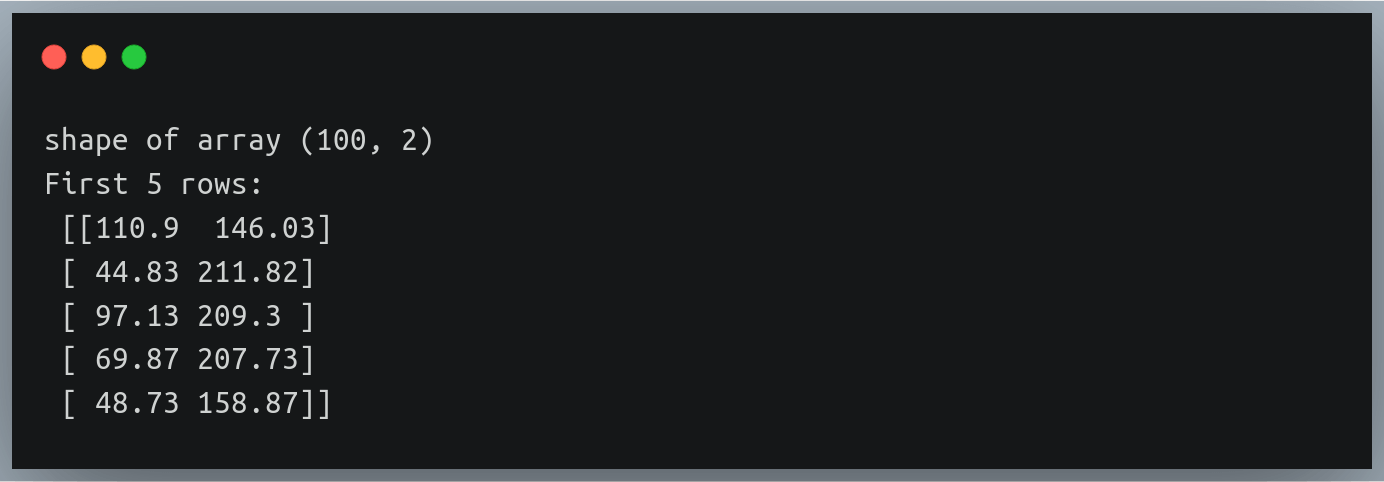
You can verify the results again by checking the shape of the data array and also printing the first few rows:
print("shape of array", data.shape)
print("First 5 rows:\n", data[:5])
Output:
Dealing with 2 delimiters
Now there may be a situation where there are more than 1 delimiters in a file.
For example, let’s imagine each of our lines contained a 3rd value representing the date of birth of the individual in dd-mm-yyyy format
110.90, 146.03, 3-7-1981
44.83, 211.82, 1-2-1986
97.13, 209.30, 14-2-1989
…
Now suppose we want to extract the dates, months and years as 3 different values into 3 different columns of our NumPy array. So should we pass “,” as the delimiter or should we pass “-”?
We can pass only 1 value to the delimiter parameter in the np.loadtxt method!
No need to worry, there is always a workaround. Let’s use a third file ‘./weight_height_3.txt’ for this example
We’ll use a naive approach first, which has the following steps:
- read the file
- eliminate one of the delimiters in each line and replace it with one common delimiter (here comma)
- append the line into a running list
- pass this list of strings to the np.loadtxt function instead of passing a file path.
Let’s write the code:
#reading each line from file and replacing "-" by ","
with open("./weight_height_3.txt") as f_input:
text = [l.replace("-", ",") for l in f_input]
#calling the loadtxt method with “,“ as delimiter
data = np.loadtxt(text, delimiter=",")
- Note that we are passing a list of strings as input and not a file path.
- When calling the function we still pass the delimiter parameter with the value “,” as we’ve replaced all instances of the second delimiter ‘-’ by a comma.
- The returned NumPy array should now have 5 columns
You can once again validate the results by printing the shape and the first five lines:
print("Shape of data:", data.shape)
print("First five rows:\n",data[:5])
Output:
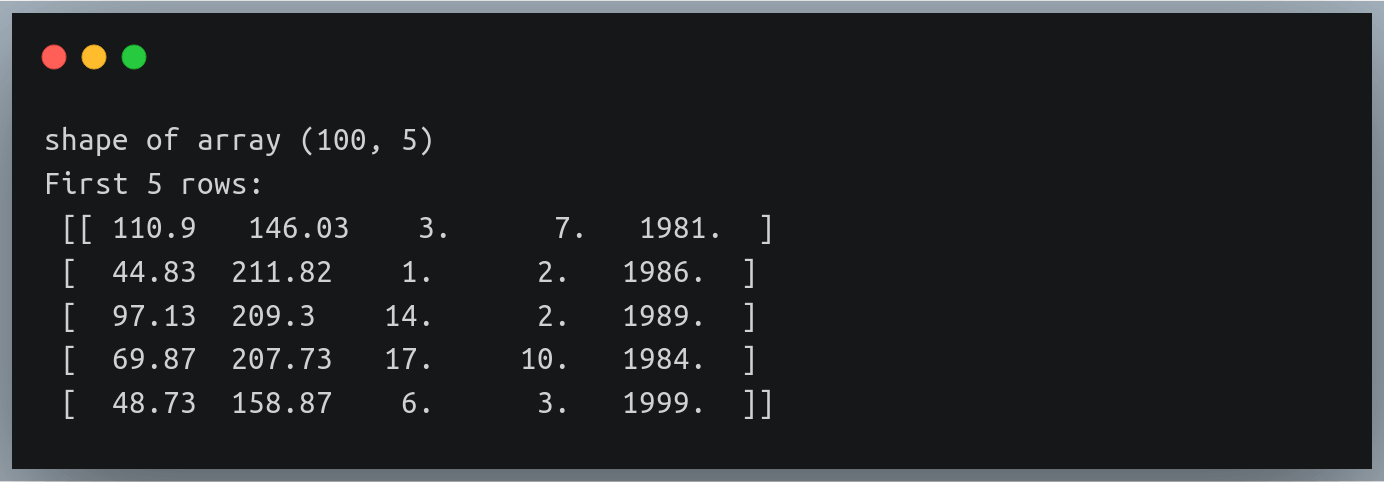
Notice how we have 3 additional columns in each row indicating the date, month and year of birth
Also notice the new values are all floating-point values; however date, month or year make more sense as integers!
We’ll look at how to handle such data type inconsistencies in the coming section.
A general approach for multiple delimiters
In this section, we will look at a general approach for working with multiple delimiters.
Also, we’ll learn how we can use generators instead of file paths – a more efficient solution for multiple delimiters, than the one we discussed in the previous section.
The problem with reading the entire file at once and storing them as a list of strings is that it doesn’t scale well. For instance, if there is a file with a million lines, storing them in a list all at once is going to consume unnecessary additional memory.
Hence we will use generators to get rid of any additional delimiter.
A generator ‘yields’ us a sequence of values on the fly i.e it will read the lines of a file as required instead of reading them all at once
So let’s first define a generator function that takes in a file path and a list of delimiters as a parameter.
def generate_lines(filePath, delimiters=[]):
with open(filePath) as f:
for line in f:
line = line.strip() #removes newline character from end
for d in delimiters:
line =line.replace(d, " ")
yield line
Here we are going through each of the delimiters one by one in each line and replacing them by a blank space ” ” which is the default delimiter in np.loadtxt function
We will now call this generator function and pass the returned generator object to the np.loadtxt method in place of the file path.
gen = generate_lines("./weight_height_3.txt", ["-",","])
data = np.loadtxt(gen)
Note that we did not need to pass any additional delimiter parameter, as our generator function replaced all instances of the delimiters in the passed list by a space, which is the default delimiter.
We can extend this idea and specify as many delimiters as needed.
Specifying the data type
Unless specified otherwise, the np.loadtxt function of the NumPy package assumes the values in the passed text file to be floating-point values by default.
So if you pass a text file that has characters other than numbers, the function will throw an error, stating it was expecting floating-point values.
We can overcome this by specifying the data type of the values in the text file using the datatypeparameter.
In the previous example, we saw the date, month and year were being interpreted as floating-point values, however, we know that these values can never exist in decimal form.
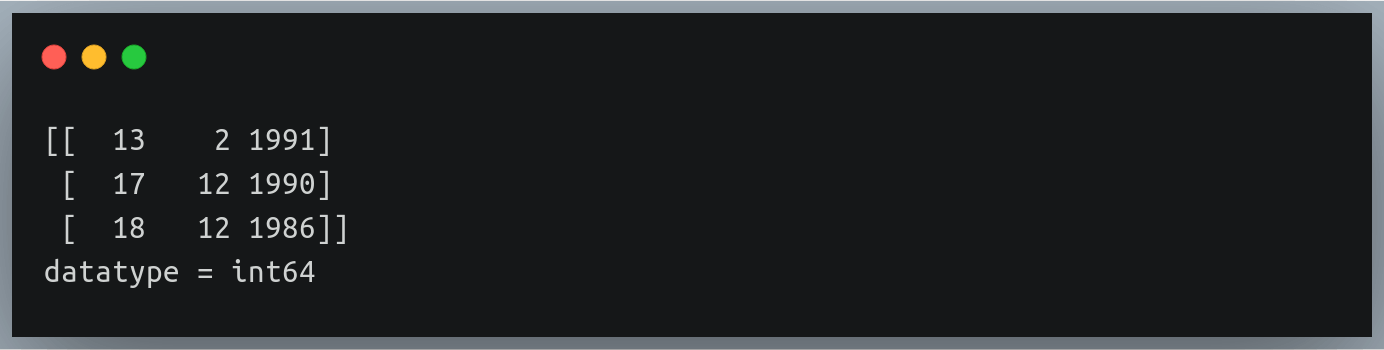
Let’s look at a new file ‘./weight_height_4.txt’ which has only 1 column for the date of birth of individuals in the dd-mm-yyyy format:
13-2-1991
17-12-1990
18-12-1986
…
So we’ll call the loadtxt method with “-” as the delimiter:
data = np.loadtxt("./weight_height_4.txt", delimiter="-")
print(data[:3])
print("datatype =",data.dtype)
If we look at the output of the above lines of code, we’ll see that each of the 3 values has been stored as floating-point values by default and the data type of the array is ‘float64’
We can alter this behavior by passing the value ‘int’ to the ‘dtype’ parameter. This will ask the function to store the extracted values as integers, and hence the data type of the array will also be int.
data = np.loadtxt("./weight_height_4.txt", delimiter="-", dtype="int")
print(data[:3])
print("datatype =",data.dtype)
Output:
But what if there are columns with different data types?
Let’s say we have the first two columns having float values and the last column having integer values.
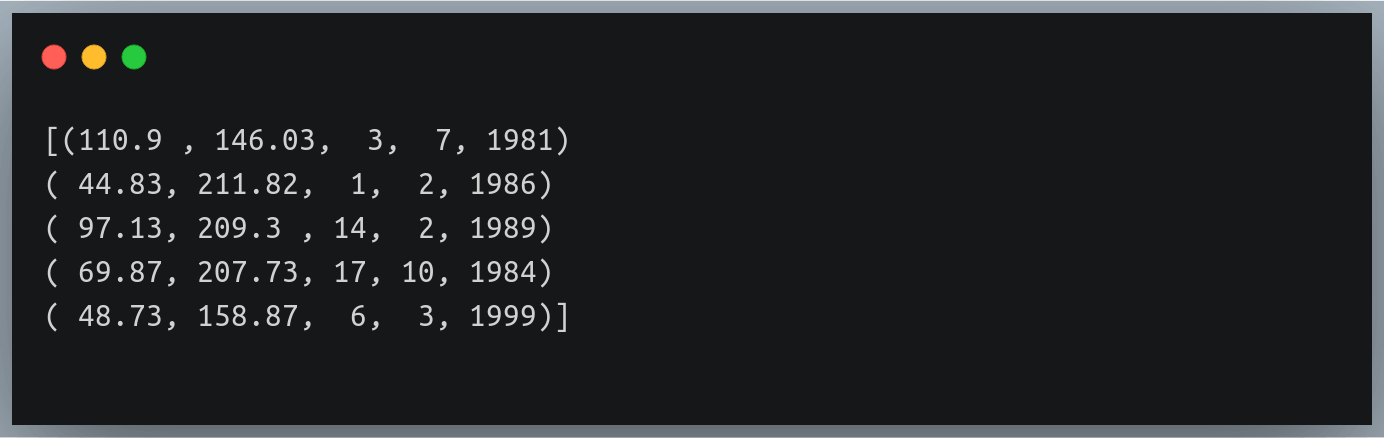
In that case, we can pass a comma-separated datatype string specifying the data type of each column (in order of their existence) to the dtype parameter.
However, in such a case the function will return a NumPy array of tuples of values since a NumPy array as a whole can have only 1 data type
Let’s try this on ‘weight_height_3.txt’ file where the first two columns (weight, height) had float values and the last 3 values (date, month, year) were integers:
Output:
Ignoring headers
In some cases (especially CSV files), the first line of the text file may have ‘headers’ describing what each column in the following rows represents. While reading data from such text files, we may want to ignore the first line because we cannot (and should not) store them in our NumPy array.
In such a case, we can use the ‘skiprows’ parameter and pass the value 1, asking the function to ignore the first 1 line(s) of the text file.
Let’s try this on a CSV file – ‘weight_height.csv’:
Weight (in Kg), Height (in cm)
73.847017017515,241.893563180437
68.7819040458903,162.310472521300
74.1101053917849,212.7408555565
…
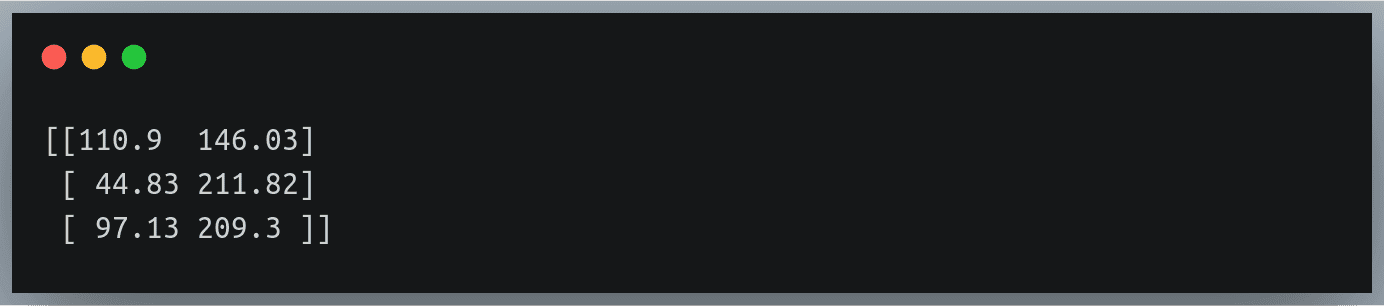
Now we want to ignore the header line i.e the first line of the file:
data = np.loadtxt("./weight_height.csv", delimiter=",", skiprows=1)
print(data[:3])
Output:
Likewise, we can pass any positive integer n to the skiprows parameter asking to ignore first n rows from the file.
Ignoring the first column
Sometimes, we may also want to skip the first column because we are not interested in it. For example, if our text file had the first column as “gender”, and if we don’t need to include the values of this column when extracting the data, we need a way to ask the function to do the same.
We do not have a skipcols parameter like skiprows in np.loadtxt function, using which, we could express this need. However, np.loadtxt has another parameter called ‘usecols’ where we specify the indices of the columns to be retained.
So if we want to skip the first column, we can simply supply the indices of all the columns except the first (remember indexing begins at zero)
Enough talking, let’s get to work!
Let’s look at the content of a new file ‘weight_height_5.txt’ which has an additional gender column that we want to ignore.
Male, 110.90, 146.03
Male, 44.83, 211.82
…
…
Female, 78.67, 158.74
Male, 105.64, 164.21
We’ll first determine the number of columns in the file from the first line and then pass a range of column indices excluding the first one:
with open("./weight_height_5.txt") as f:
#determining number of columns from the first line of text
n_cols = len(f.readline().split(","))
data = np.loadtxt("./weight_height_5.txt", delimiter=",",usecols=np.arange(1, n_cols))
print("First five rows:\n",data[:5])
Here we are supplying a range of values beginning from 1 (second column) and extending up to n_cols (the last column)
Output:

We can generalize the use of the usecols parameter by passing a list of indices of only those columns that we want to retain.
Load first n rows
Just as we can skip the first n rows using the skiprows parameter, we can also choose to load only the first n rows and skip the rest. This can be achieved using the max_rows parameter of the np.loadtxt method.
Let us suppose that we want to read only the first 10 rows from the text file ‘weight_height_2.txt’. We’ll call the np.loadtxt method along with the max_rows parameter and pass the value 10.
data = np.loadtxt("./weight_height_2.txt", delimiter=",",max_rows = 10)
print("Shape of data:",data.shape)
Output:

As we can see, the returned NumPy array has only 10 rows which are the first 10 rows of the text file.
If we use the max_rows parameter along with skiprowsparameter, then the specified number of rows will be skipped and next n rows will be extracted where n is the value we pass to max_rows.
Load specific rows
If we want the np.loadtxt function to load only specific rows from the text file, no parameter supports this feature.
However, we can achieve this by defining a generator that accepts row indices and returns lines at those indices. We’ll then pass this generator object to our np.loadtxt method.
Let’s first define the generator:
def generate_specific_rows(filePath, row_indices=[]):
with open(filePath) as f:
# using enumerate to track line no.
for i, line in enumerate(f):
#if line no. is in the row index list, then return that line
if i in row_indices:
yield line
Let’s now use the np.loadtxt function to read the 2nd, 4th and 100th line in the file ‘weight_height_2.txt’
gen = generate_specific_rows("./weight_height_2.txt",row_indices = [1, 3, 99])
data = np.loadtxt(gen, delimiter=",")
print(data)
This should return a NumPy array having 3 rows and 2 columns:
Skip the last row
If you want to exclude the last line of the text file, you can achieve this in multiple ways. You can either define another generator that yields lines one by one and stops right before the last one, or you can use an even simpler approach – just figure out the number of lines in the file, and pass 1 less than that count to the max_rows parameter.
But how will you figure out the number of lines?
Follow along!
with open("./weight_height_2.txt") as f:
n = len(list(f))
print("n =", n)
Now n contains the number of lines present in `weight_height_2.txt` file, that value should be 100.
We will now read the text file as we used to, using the np.loadtxt method along with the max_rows parameter with value n – 1.
data = np.loadtxt("./weight_height_2.txt", delimiter=",",max_rows=n - 1)
print("data shape =",data.shape)
Output:

As we can see, the original text file had 100 rows, but when we read data from the file, it’s shape is (99, 2) since it skipped the last row from the file.
Skip specific columns
Suppose you wanted to ignore some of the columns while loading data from a text file by specifying the indices of such columns.
While the np.loadtxt method provides a parameter to specify which columns to retain (usecols), it doesn’t offer a way to do the opposite i.e specify which columns to skip. However, we can always find a workaround!
We shall first define the indices of columns to be ignored, and then using them we will derive the list of indices to be retained as the two sets would be mutually exclusive.
We will then pass this derived indices list to the usecols parameter.
Here is pseudocode for the entire process:
- Find the number of columns in the file n_cols (explained in an earlier section)
- Define the list of indices to be ignored
- Create a range of indices from 0 to n_cols, and eliminate the indices of step 2 from this range
- Pass this new list to usecols parameter in np.loadtxt method
Let’s create a wrapper function loadtext_without_columns that implements all the above steps:
def loadtext_without_columns(filePath, skipcols=[], delimiter=","):
with open(filePath) as f:
n_cols = len(f.readline().split(delimiter))
#define a range from 0 to n_cols
usecols = np.arange(0, n_cols)
#remove the indices found in skipcols
usecols = set(usecols) - set(skipcols)
#sort the new indices in ascending order
usecols = sorted(usecols)
#load the file and retain indices found in usecols
data = np.loadtxt(filePath, delimiter = delimiter, usecols = usecols)
return data
To test our code, we will work with a new file `weight_height_6.txt` which has 5 columns – the first two columns indicate width and height and the remaining 3 indicate the date, month and year of birth of the individuals.
All the values are separated by a single delimiter – comma:
110.90, 146.03, 3,7,1981
44.83, 211.82, 1,2,1986
97.13, 209.30, 14,2,1989
…
…
105.64, 164.21, 3,6,2000
Suppose we were not interested in the height and the date of birth of the individual, and so we wanted to skip the columns at positions 1 and 2.
Let’s call our wrapper function specifying our requirements:
data = loadtext_without_columns("./weight_height_6.txt",skipcols = [1, 2], delimiter = ",")
# print first 5 rows
print(data[:5])
Output:

We can see that our wrapper function only returns 3 columns – weight, month and year. It has ensured that the columns we specified have been skipped!
Load 3D arrays
So far we’ve been reading the contents of the file as a 2D NumPy array. This is the default behavior of the np.loadtxt method, and there’s no additional parameter that we can specify to interpret the read data as a 3D array.
So the simplest approach to solve this problem would be to read the data as a NumPy array and use NumPy’s reshape method to reshape the data in any shape of any dimension that we desire.
We just need to be careful that if we want to interpret it as a multidimensional array, we should make sure it is stored in the text file in an appropriate manner and that after reshaping the array, we’d get what we actually desired.
Let us take an example file – ‘weight_height_7.txt’.
This is the same file as ‘weight_height_2.txt’. The only difference is that this file has 90 rows, and each 30-row block represents a different section or class to which individuals belong.
So there are a total of 3 sections (A, B and C) – each having 30 individuals whose weights and heights are listed on a new row.
The section names are denoted with a comment just before the beginning of each section (you can check this at lines 1, 32 and 63).
The comment statements begin with ‘#’ and these lines are ignored by np.loadtxt when reading the data. We can also specify any other identifier for comment lines using the parameter ‘comments’
Now when you read this file, and print its shape, it would display (90,2) because that is how np.loadtxt reads the data – it arranges a multi-row data into 2D arrays.
But we know that there is a logical separation between each group of 30 individuals, and we would want the shape to be (3, 30, 2) – where the first dimension indicates the sections, the second one represents each of the individuals in that section and the last dimension indicates the number of values associated to each of these individuals (here 2 for weight & height).
Using NumPy reshape method
So we want our data to be represented as a 3D array.
We can achieve this by simply reshaping the returned data using NumPy’s reshape method.
data = np.loadtxt("./weight_height_7.txt",delimiter=",")
print("Current shape = ",data.shape)
Settingsdata = data.reshape(3,30,2)
print("Modified shape = ",data.shape)
print("fifth individual of section B - weight, height =",data[1,4,:])
Output:
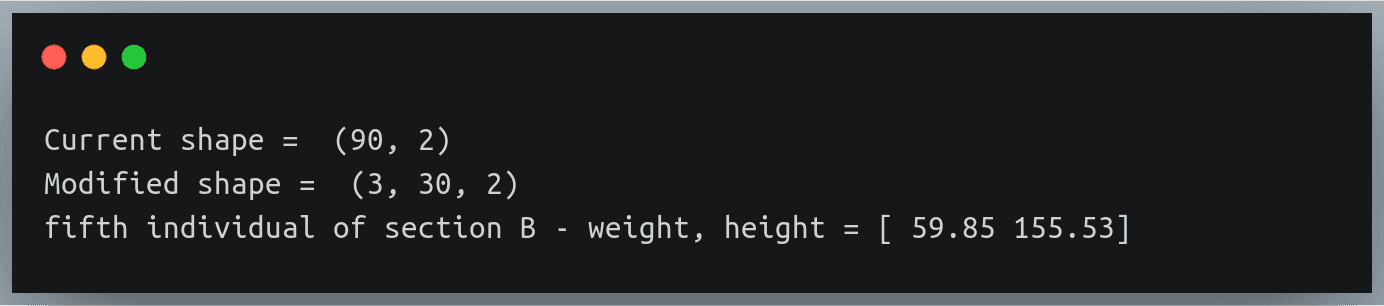
Notice how we are printing the details of a specific individual using 3 indices
The returned result belongs to the 5th individual of section B – this can be validated from the text:
…
#section B
100.91, 155.55
72.93, 150.38
116.68, 137.15
86.51, 172.15
59.85, 155.53
…
Comparison with alternatives
While numpy.loadtxt is an extremely useful utility for reading data from text files, it is not the only one!
There are many alternatives out there that can do the same task as np.loadtxt, many of these are better than np.loadtxt in many aspects. Let’s briefly look at 3 such alternative functions.
numpy.genfromtxt
- This is the most discussed and the most used method alongside np.loadtxt
- There’s no major difference between the two, the only one that stands out is np.genfromtxt’s ability to smoothly handle missing values.
- In fact, NumPy’s documentation describes np.loadtxt as “an equivalent function (to np.genfromtxt) when no data is missing.
- So the two are almost similar methods, except that np.genfromtxt can do more sophisticated processing of the data in a text file.
numpy.fromfile
- np.fromfile is commonly used when working with data stored in binary files, with no delimiters.
- It can read plain text files but does so with a lot of issues (go ahead and try reading the files we discussed using np.fromfile)
- While it is faster in execution time than np.loadtxt, but it is generally not a preferred choice when working with well-structured data in a text file.
- Besides NumPy’s documentation mentions np.loadtxt as a ‘more flexible (than np.fromfile) way of loading data from a text file.
pandas.read_csv
- pandas.read_csv is the most popular choice of Data Scientists, ML Engineers, Data Analysts, etc. for reading data from text files.
- It offers way more flexibility than np.loadtxt or np.genfromtxt.
- Although you cannot pass a generator to pandas.read_csv as we did.
- In terms of speed of execution, however, pandas.read_csv do better than np.loadtxt
Handling Missing Values
As discussed in our section comparing np.loadtxt with other options, np.genfromtxt handles missing values by default. We do not have any direct way of handling missing values in np.loadtxt
Here we’ll look at an indirect (and a slightly sophisticated) way of handling missing values with the np.loadtxt method.
The converters parameter:
- np.loadtxt has a converters parameter that is used to specify the preprocessing (if any) required for each of the columns in the file.
- For example, if the text file stores the height column in centimeters and we want to store them as inches, we can define a converter for the heights column.
- The converters parameter accepts a dictionary where the keys are column indices and the values are methods that accept the column value, ‘convert’ it and return the modified value.
How can we use converters to handle missing values?
- We need to first decide the default datatype i.e the value to be used to fill in the positions where the actual values are missing. Let’s say we want to fill in the missing height and weight values with 0, so our fill_value will be 0.
- Next, we can define a converter for each column in the file, which checks if there is some value or an empty string in that column and if it’s an empty string, it will fill it with our fill_value.
- To do this, we’ll have to find the number of columns in the text file, and we have already discussed how to achieve this in an earlier section.
We’ll use the file ‘weight_height_8.txt’ which is the same as ‘weight_height_2.txt’ but has several missing values.
, 146.03
44.83, 211.82
97.13,
69.87, 207.73
, 158.87
99.25, 195.41
…
Let’s write the code to fill in these missing values’ positions with 0.
# finding number of columns in the file
with open("./weight_height_8.txt") as f:
n_cols = len(f.readline().split(","))
print("Number of columns", n_cols)
# defining converters for each of the column (using 'dictionary
# comprehension') to fill each missing value with fill_value
fill_value = 0
converters = {i: lambda s: float(s.strip() or fill_value) for i in range(2)}
data = np.loadtxt("./weight_height_8.txt", delimiter=",",converters = converters)
print("data shape =",data.shape)
print("First 5 rows:\n",data[:5])
Output:
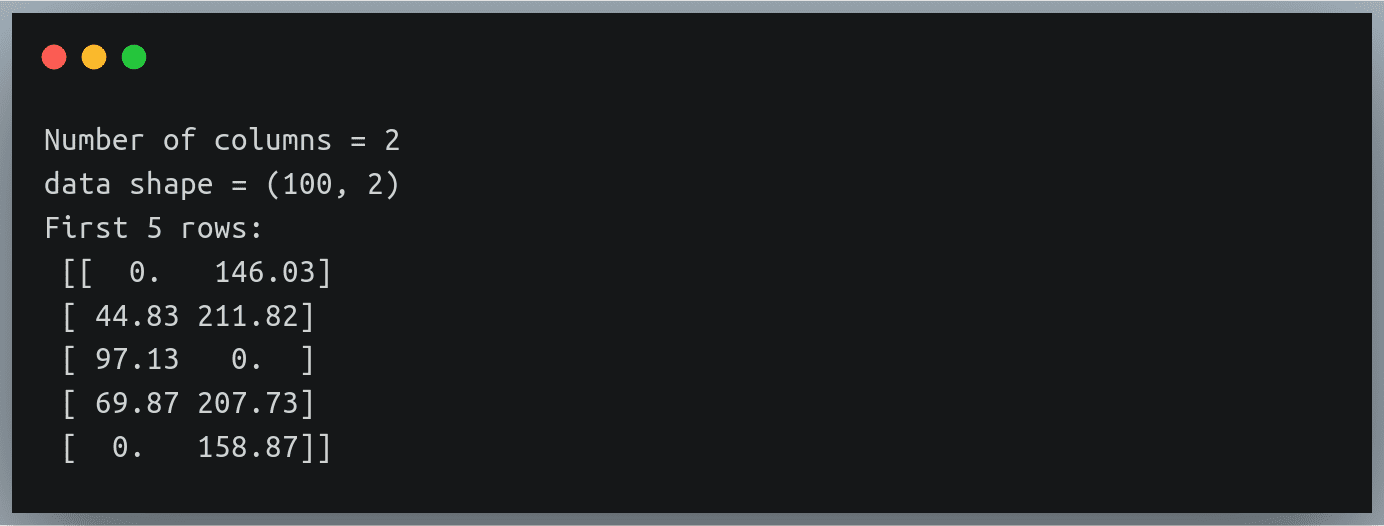
The missing height and weight values have been correctly replaced with a 0. No magic!
Conclusion
numpy.loadtxt is undoubtedly one of the most standard choices for reading a well-structured data stored in a text file. It offers us great flexibility in choosing various options for specifying the way we want to read the data, and wherever it doesn’t – remember there’s always a workaround!



 In this post, we will convert an array to object using
In this post, we will convert an array to object using